Beautiful Correlation Tables in R
I have achieved another victory in getting R to produce SPSS-like results. In experimental psychology, an analysis of measurement variable correlations is a common method in the course of a statistical analysis. Thus, I wanted R to produce a publication-quality output similar to SPSS: a correlation matrix of measurement variables that contains only the lower triangle of observations, where observations have two decimal digits and are flagged with stars (*, **, and ***) according to levels of statistical significance. However, as statmethods notices:
> corstarsl(swiss[,1:4])
Fertility Agriculture Examination
Fertility
Agriculture 0.35*
Examination -0.65*** -0.69***
Education -0.66*** -0.64*** 0.70***
If one employs the xtable package that produces LaTeX tables from within R, xtable(corstarsl(swiss[,1:4])) produces this:
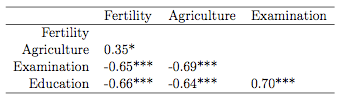 Isn't that beautiful? I like it a lot. Here's the code (as I said, much of it taken from here):
Isn't that beautiful? I like it a lot. Here's the code (as I said, much of it taken from here):
corstarsl <- function(x){
require(Hmisc)
x <- as.matrix(x)
R <- rcorr(x)$r
p <- rcorr(x)$P
## define notions for significance levels; spacing is important.
mystars <- ifelse(p < .001, "***", ifelse(p < .01, "** ", ifelse(p < .05, "* ", " ")))
## trunctuate the matrix that holds the correlations to two decimal
R <- format(round(cbind(rep(-1.11, ncol(x)), R), 2))[,-1]
## build a new matrix that includes the correlations with their apropriate stars
Rnew <- matrix(paste(R, mystars, sep=""), ncol=ncol(x))
diag(Rnew) <- paste(diag(R), " ", sep="")
rownames(Rnew) <- colnames(x)
colnames(Rnew) <- paste(colnames(x), "", sep="")
## remove upper triangle
Rnew <- as.matrix(Rnew)
Rnew[upper.tri(Rnew, diag = TRUE)] <- ""
Rnew <- as.data.frame(Rnew)
## remove last column and return the matrix (which is now a data frame)
Rnew <- cbind(Rnew[1:length(Rnew)-1])
return(Rnew)
}
Unfortunately, neither cor( ) or cov( ) produce tests of significance, although you can use the cor.test( ) function to test a single correlation coefficient.I did a little research and found this post on the R-help list. I modified Chuck Cleland's code a little so that the following command on the swiss data frame (provided in the Hmisc package) produces a beautiful output:
> corstarsl(swiss[,1:4])
Fertility Agriculture Examination
Fertility
Agriculture 0.35*
Examination -0.65*** -0.69***
Education -0.66*** -0.64*** 0.70***
If one employs the xtable package that produces LaTeX tables from within R, xtable(corstarsl(swiss[,1:4])) produces this:
 Isn't that beautiful? I like it a lot. Here's the code (as I said, much of it taken from here):
Isn't that beautiful? I like it a lot. Here's the code (as I said, much of it taken from here):corstarsl <- function(x){
require(Hmisc)
x <- as.matrix(x)
R <- rcorr(x)$r
p <- rcorr(x)$P
## define notions for significance levels; spacing is important.
mystars <- ifelse(p < .001, "***", ifelse(p < .01, "** ", ifelse(p < .05, "* ", " ")))
## trunctuate the matrix that holds the correlations to two decimal
R <- format(round(cbind(rep(-1.11, ncol(x)), R), 2))[,-1]
## build a new matrix that includes the correlations with their apropriate stars
Rnew <- matrix(paste(R, mystars, sep=""), ncol=ncol(x))
diag(Rnew) <- paste(diag(R), " ", sep="")
rownames(Rnew) <- colnames(x)
colnames(Rnew) <- paste(colnames(x), "", sep="")
## remove upper triangle
Rnew <- as.matrix(Rnew)
Rnew[upper.tri(Rnew, diag = TRUE)] <- ""
Rnew <- as.data.frame(Rnew)
## remove last column and return the matrix (which is now a data frame)
Rnew <- cbind(Rnew[1:length(Rnew)-1])
return(Rnew)
}
Labels: psychology, R, spss, statistics

